Two years ago this month, the Nobel Prize in Chemistry was awarded to two of the pioneers of the Crispr DNA-editing technique. Since then, it has displaced all other laboratory techniques for that purpose by virtue of its accuracy, simplicity, ease of use, and low cost.
How it works and why it was Nobel-worthy

Image provided by the authors
To use Crispr, you start by creating a target/guide string of ~20 RNA bases that are complementary to the location where you want to delete or modify a gene. You combine that with an enzyme (the original enzyme used in Crispr is an endonuclease called Cas9), mix the two with DNA, and the Crispr/Cas9 complex will bind to that exact location and edit it for you. (Of course, in practice it is a little more complex, but not much).
Image provided by the authors
Until surprisingly recently, gene “editing” was accomplished by using radiation or chemicals to randomly mutate DNA (e.g., in thousands of fruit flies), then looking at all of the results to see what happened (red or white fruit fly eyes, anyone?). More recently, improved but complex mutation tools were developed, such as PCR-directed mutation and enzymatic techniques (Zinc Finger Nucleases – ZFNs, or TALENS). All of these have been entirely obsoleted by Crispr.
How Crispr diagnostics work
The pandemic accelerated research into the diagnostic uses of Crispr dramatically. Although just three Crispr-based SARS-CoV-2 tests are authorized (two versions of DETECTR from Mammoth Biosciences, plus one from Sherlock) a recent review summarized 36 novel Crispr techniques in development for diagnostics and that was just a subset – with names like MeCas12a, CREST, iSCAN, CASdetec, FELUDA, RADICA, ENHANCE, IMPACT, CASMEAN, CASCADE, SHINE, and SATORI. Combining Crispr with the enormous progress in microfluidics and lab-on-a-chip offers an exciting path forward.
A Crispr complex will only attach when it finds the unique stretch of the viral genome to which it was targeted (works for bacteria, amoeba, and fungi genomes, too), which is analogous to what PCR primers do. But PCR then does something Crispr cannot: once it finds its target, it generates billions of copies, the presence of which is easy to detect. Many copies means the sample is positive; very few copies means negative.
The RTqPCR diagnostic process involves 4 basic steps: collect/neutralize sample; purify/extract RNA; convert to DNA (the “RT” part); perform automated quantified PCR (the “q” part reveals viral load). PCR itself is a complex process – it requires temperature cycling: heat to ~95°C (separate strands), cool to 60°C (attach primers), heat to 70°C (create copies), add fluorescent probes (for the “q”), repeat ~40 times. As a result, simpler isothermal (~65°C) techniques were developed, and are used by most COVID molecular POC tests, at some sacrifice of fidelity. All the steps require careful handling – moving samples between machines; adding different reagents at different times and at different temperatures, all without contamination.
Crispr diagnostics basically require all these steps plus one: the addition of Crispr towards the end.
The big strength of Crispr is that its fluorescent probes are not generic – they only report when the Crispr/Cas complex binds to its correct targets – but if you must do PCR anyway, is it worth the bother to do Crispr as well? As always, the devil is in the details, but one thing Crispr diagnostics do provide is double protection against some common PCR errors. If PCR amplifies the wrong target, Crispr will not be fooled.
All new diagnostics need fine-tuning, Crispr not excepted.
Forty years before Crispr, chemistry Nobel winner (and surfer) Kary Mullis sketched the concept of PCR on a napkin, but until a heat-tolerant polymerase was identified from a Yellowstone hot spring, PCR was labor-intensive and slow. Crispr diagnostics have just begun this fine-tuning process, 38 years behind PCR, but enormous progress is being made.
The most striking example of Crispr progress is the discovery of novel Cas enzymes which expand on Cas9’s properties: Cas12a (Mammoth) and Cas13a (Sherlock, illustrated below) find target DNA the way Cas9 does, but then they also cut any nearby single strand DNA (ssDNA). If you include short ssDNA strands attached to quenched fluorescent reporter molecules, they get cleaved too, releasing fluorescence once binding has occurred. (Cas12a binds DNA, while Cas13a can bind RNA directly without it being converted to DNA first.)
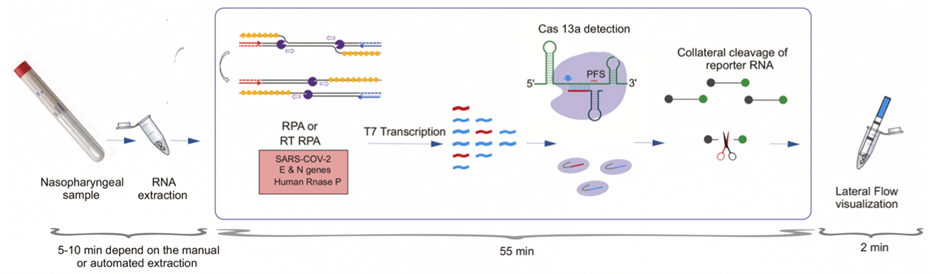
Image provided by the authors
The traditional way to deal with all this is to employ skilled technicians, in a high throughput sterile lab, and all three currently authorized Crispr COVID tests go this way. However, there is emerging wave of low throughput microfluidic chips combined with simple instruments (handheld or desktop, even a mobile phone) the future for POC.
Nevertheless, Crispr’s need for prior purification and amplification (either PCR or isothermal) remains the single biggest challenge for the technique’s use in diagnostics. Many clever ideas to deal with the problem are being developed, but none have yet overcome (or minimized) this duplication disadvantage versus PCR.
Source: Sensitive & Specific: The Testing Newsletter
Authors: Mara G. Aspinall and Liz Ruark
Republished with Permission
